Coding
(in Stata and R)
2025-02-14
Overview
In this session we’ll cover:
- R fundamentals
- Common coding tasks
- Concept
- Data
- Code (Stata and R)
R fundamentals
R and RStudio
R is the language that the code is written in.
RStudio is the software many people use to write R code in.
At a minimum you need to install R (r-project.org)
IDE options include:
- RStudio (posit.co/products/open-source/rstudio)
- Positron (positron.posit.co)
- VS Code (code.visualstudio.com)
Packages
One of the greatest strengths of R is that it is open-source and there are an enormous number of packages available.
A package is a collection of functions usually written around a particular goal or task.
Packages I recommend:
tidyversewhich includes:dplyrfor data manipulationggplot2for data visualisationstringrfor working with text datalubridatefor working with dates
janitorwhich has many useful cleaning toolshavenfor reading Stata files
Packages
First you need to install packages:
Then you need to load them into your session using library():
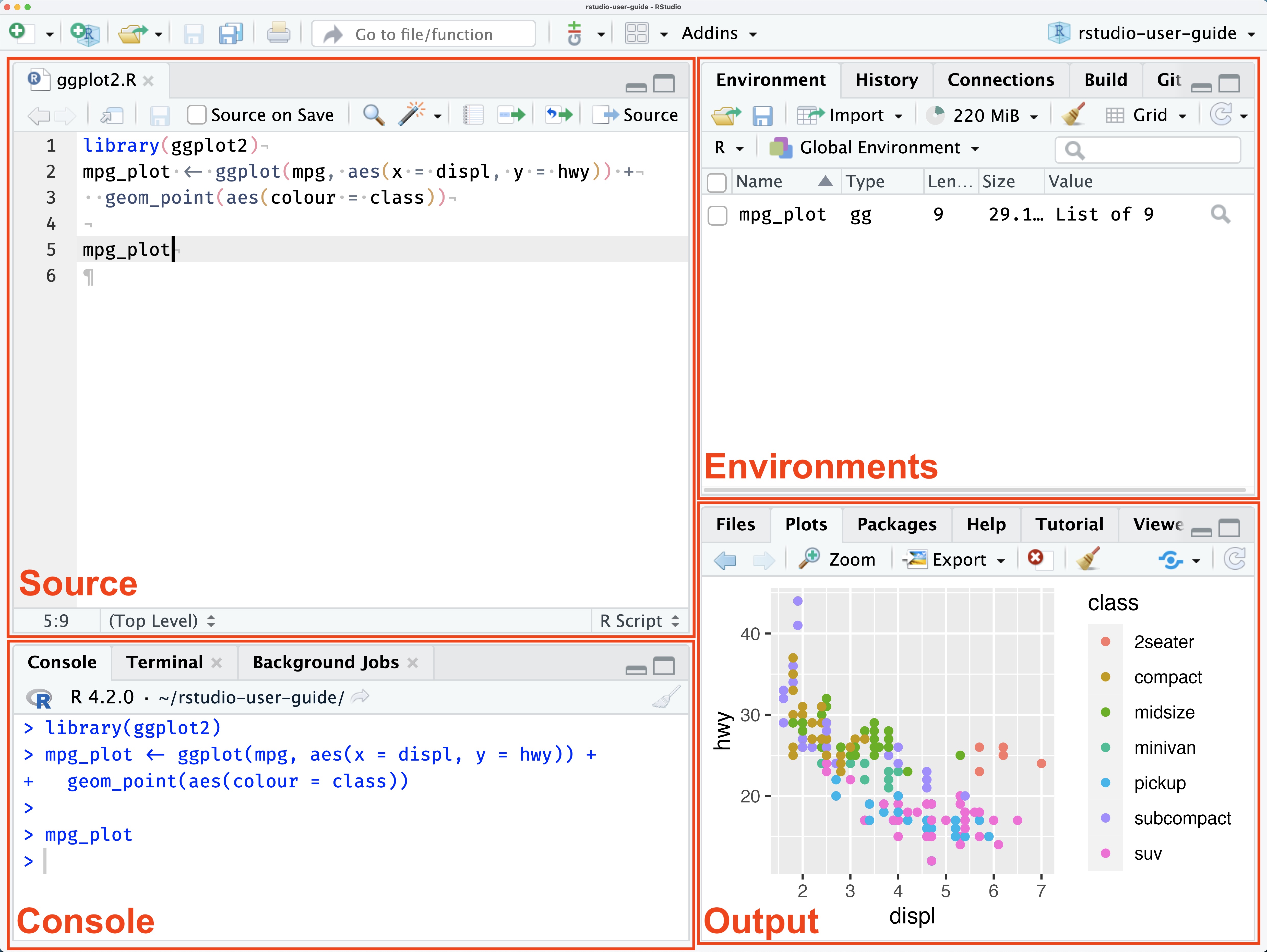
Assignment
In Stata, you load one dataset and all commands are executed in relation to the currently loaded dataset.
In R, the default is for commands to output their result in the console.
This means you need to assign the output of your commands to something if you want to store it.

Assignment
If we read in a dataset without assigning anything, it just gets displayed:
# A tibble: 74 × 12
make price mpg rep78 headroom trunk weight length turn displacement
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 AMC Concord 4099 22 3 2.5 11 2930 186 40 121
2 AMC Pacer 4749 17 3 3 11 3350 173 40 258
3 AMC Spirit 3799 22 NA 3 12 2640 168 35 121
4 Buick Cent… 4816 20 3 4.5 16 3250 196 40 196
5 Buick Elec… 7827 15 4 4 20 4080 222 43 350
6 Buick LeSa… 5788 18 3 4 21 3670 218 43 231
7 Buick Opel 4453 26 NA 3 10 2230 170 34 304
8 Buick Regal 5189 20 3 2 16 3280 200 42 196
9 Buick Rivi… 10372 16 3 3.5 17 3880 207 43 231
10 Buick Skyl… 4082 19 3 3.5 13 3400 200 42 231
# ℹ 64 more rows
# ℹ 2 more variables: gear_ratio <dbl>, foreign <dbl+lbl>Instead we have to give it a name and assign to the environment:
Multiple datasets
Being able to have many things assigned in the environment at once opens up many possibilities:
- Working on multiple datasets simultaneously
- Easier than working with Stata’s
frame
- Easier than working with Stata’s
- Creating subsets of data
- More powerful version of Stata’s
preserveandrestore
- More powerful version of Stata’s
- Outputs from multiple analyses are readily available
- Model results, data visualisations
Pipes
In Stata, you write lines of code that get executed one after the other, individually and sequentially.
In R, you can create pipelines using pipes (|>) to link functions together and avoid having to repeatedly refer to the data you want to work with.
Pipes
Without pipes
First step: Get the data
Reading in a data file
The data you want to work with is contained in a file located somewhere.
To start using the data you need to consider:
- The location of the file
- Is the data on your computer? A shared drive? An online location?
- Are you using the absolute or relative pathway?
- How are others going to run your code?
- The format of the file (
.dta,.xlsx,.csv,.txt,.SAV)
A note on folder structure
Relative vs. absolute directories
Users/Ben/Documents/GitHub/workshops/2025-02-14-coding-in-stata-and-r/data/auto.dta./data/auto.dta
Network locations, root location different according to machine type.
Generally speaking, avoid using absolute pathways when possible.
Navigate to other directories from the current working directory using .. to go ‘up’ a folder:
../2024-08-20-admin-data/img/aihw.PNG
A note on directories
In Stata you would set your working directory using cd, such as
In R you can use setwd() to set your working directory
I recommend using R Projects instead - note the .Rproj file below. Opening this file will set the directory automatically to the location of that file.
- 2025-02-14-coding-in-stata-and-r
- _brand.yml
- 2025-02-14-coding-in-stata-and-r.Rproj
- slides.html
- slides.qmd
- data
- imgReading in a data file
For a file in the same location as the current working directory:
For a file that is in another folder:
For a file that is online:
For a file in the same location as the current working directory:
For a file that is in another folder:
For a file that is online:
Overview of the data
Raw data containing observations of three different types of penguins, made available through https://allisonhorst.github.io/palmerpenguins/
| studyName | Sample Number | Species | Region | Island | Stage | Individual ID | Clutch Completion | Date Egg | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAL0910 | 66 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N99A2 | No | 2009-11-21 | 49.6 | 18.2 | 193 | 3775 | MALE | 9.46180 | -24.70615 | Nest never observed with full clutch. |
| PAL0910 | 104 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N23A2 | Yes | 2009-11-18 | 49.1 | 15.0 | 228 | 5500 | MALE | 8.65914 | -25.79203 | NA |
| PAL0910 | 123 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N43A1 | Yes | 2009-11-22 | 45.2 | 14.8 | 212 | 5200 | FEMALE | 8.24246 | -26.11969 | NA |
| PAL0708 | 23 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N42A1 | Yes | 2007-11-27 | 46.5 | 14.5 | 213 | 4400 | FEMALE | 7.90436 | -25.39470 | NA |
| PAL0708 | 22 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N73A2 | Yes | 2007-12-03 | 48.5 | 17.5 | 191 | 3400 | MALE | 9.42666 | -24.26375 | NA |
| PAL0708 | 25 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N44A1 | Yes | 2007-11-29 | 42.9 | 13.1 | 215 | 5000 | FEMALE | 7.68528 | -25.39181 | NA |
| PAL0809 | 80 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N60A2 | Yes | 2008-11-09 | 45.2 | 16.4 | 223 | 5950 | MALE | 8.19749 | -26.65931 | NA |
| PAL0809 | 52 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N13A2 | Yes | 2008-11-04 | 48.5 | 14.1 | 220 | 5300 | MALE | 8.39867 | -26.79358 | NA |
| PAL0708 | 25 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N89A1 | No | 2007-11-28 | 46.7 | 17.9 | 195 | 3300 | FEMALE | 9.74144 | -24.59467 | Nest never observed with full clutch. |
| PAL0809 | 43 | Gentoo penguin (Pygoscelis papua) | Anvers | Biscoe | Adult, 1 Egg Stage | N7A1 | Yes | 2008-11-04 | 45.3 | 13.7 | 210 | 4300 | FEMALE | 8.37615 | -26.72791 | NA |
Second step: Prepare the data
Renaming variables
Variables might be imported with difficult to work with names, so we want to rename them
# A tibble: 6 × 17
studyName `Sample Number` Species Region Island Stage `Individual ID`
<chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 PAL0708 1 Adelie Penguin … Anvers Torge… Adul… N1A1
2 PAL0708 2 Adelie Penguin … Anvers Torge… Adul… N1A2
3 PAL0708 3 Adelie Penguin … Anvers Torge… Adul… N2A1
4 PAL0708 4 Adelie Penguin … Anvers Torge… Adul… N2A2
5 PAL0708 5 Adelie Penguin … Anvers Torge… Adul… N3A1
6 PAL0708 6 Adelie Penguin … Anvers Torge… Adul… N3A2
# ℹ 10 more variables: `Clutch Completion` <chr>, `Date Egg` <date>,
# `Culmen Length (mm)` <dbl>, `Culmen Depth (mm)` <dbl>,
# `Flipper Length (mm)` <dbl>, `Body Mass (g)` <dbl>, Sex <chr>,
# `Delta 15 N (o/oo)` <dbl>, `Delta 13 C (o/oo)` <dbl>, Comments <chr>Stata automatically renames variables that contain spaces and other invalid characters, R does not.
Renaming variables
In Stata, the old name comes first, the new name comes second
Like saying “replace old name with this new name”
Renaming variables
The janitor package contains a handy function called clean_names():
[1] "study_name" "sample_number" "species"
[4] "region" "island" "stage"
[7] "individual_id" "clutch_completion" "date_egg"
[10] "culmen_length_mm" "culmen_depth_mm" "flipper_length_mm"
[13] "body_mass_g" "sex" "delta_15_n_o_oo"
[16] "delta_13_c_o_oo" "comments" Dropping/keeping variables
To keep only the specified variables, we use keep.
To drop the specified variables, we use drop.
To keep only the specified variables, we use select() from the dplyr package.
To drop the specified variables, we add a minus (-) before the name of the variable.
We can also select ranges:
tidyselect provides lots of helpful functions for use in select()!
everything() selects every variable, so the below puts study_name and individual_id first, followed by all other variables in their current order
You can select variables that have names matching a pattern. starts_with() and ends_with() will match the pattern to the start or end of the variable name
matches() looks for it anywhere
Dropping/keeping observations
Observations (rows) are also dropped using drop.
Rows are are dropped using filter().
Only rows that meet the condition are kept, use the ‘not’ operator (!) instead of drop and keep.
Note that to drop missing values you must use is.na(), you cannot say samplenumber != NA. NA is a special class in R.
filter() also comes from the dplyr package.
Dropping/keeping observations
Multiple conditions are specified with commas, where all must be true:
You can also make use of %in% with lists to make cleaner criteria:
Creating or modifying variables
Several options in Stata:
genfor making new variablesegenfor new variables based on a specific functionreplacefor modifying existing variablesclonevarfor copying existing variables
Conditions
The if argument is where conditions are typically specified in Stata.
Common ways of specifying conditions in R:
ifelse(), in base Rif_else()andcase_when()fromdplyr
Missing
Display
Stata displays missing values in strings as blank "", and uses . in all other cases.
R displays all missing values as NA, meaning the empty string "" is not considered missing.
Interpretation
Stata interprets . as a large positive number, which can cause issues in conditional statements.
R does not do this, missing is a separate category.
Variable types
Generally speaking, in Stata everything is either a number or a string.
Numbers can be bytes, integers, decimals, floats, doubles.
Categorical data can be represented as a string or as a label on a numeric variable.
Logical variables (TRUE or FALSE) do not exist.
R has more data types than Stata, including logical variables - click on the other tabs to learn more.
In the example below, values is a vector that’s a mix of numbers and strings (characters). The strings are in quotes, the numbers are not.
[1] "-4" "0" "1" "1" "2.5" "05" "11.7" "T" "FALSE"[1] -4 0 1 1 2 5 11 NA NANotice that using as.integer() truncates 2.5 and 11.7, it does not round them.
[1] "-4" "0" "1" "1" "2.5" "05" "11.7" "T" "FALSE"[1] -4.0 0.0 1.0 1.0 2.5 5.0 11.7 NA NA[1] -4 0 1 1 2 5 12 NA NANotice that round() rounds decimals of .5 toward the nearest even number .
Use round_half_up() from the janitor package to round up.
[1] "-4" "0" "1" "1" "2.5" "05" "11.7" "T" "FALSE"[1] NA NA NA NA NA NA NA TRUE FALSEStrings with T/TRUE/F/FALSE are converted to TRUE and FALSE
If we convert them to numbers first, any numbers that are not 0 are now TRUE and 0 is FALSE.
The converse is true as well - you can sum logicals as they are treated like 1 and 0
Jumping back to the penguins data, categorical data can be stored as factors:
If we tabulate the character version island, it’s sorted alphabetically
data$island n percent
Biscoe 168 0.4883721
Dream 124 0.3604651
Torgersen 52 0.1511628When we use the factor version, the result is ordered by the levels we specified.
Reshaping longer
The tidyverse package makes this very easy with pivot_longer().
data |>
select(individual_id, region, stage) |>
pivot_longer(
cols = c(region, stage),
names_to = "variable",
values_to = "number"
)# A tibble: 688 × 3
individual_id variable number
<chr> <chr> <chr>
1 N1A1 region Anvers
2 N1A1 stage Adult, 1 Egg Stage
3 N1A2 region Anvers
4 N1A2 stage Adult, 1 Egg Stage
5 N2A1 region Anvers
6 N2A1 stage Adult, 1 Egg Stage
7 N2A2 region Anvers
8 N2A2 stage Adult, 1 Egg Stage
9 N3A1 region Anvers
10 N3A1 stage Adult, 1 Egg Stage
# ℹ 678 more rowscols = specifies which columns you want reshape
names_to = names the column that the variable names will be put in
values_to = names the column that the values of those variables will be put in
Reshaping wider
As you might’ve guessed, pivot_wider() does the opposite
This time we don’t specify cols
names_from = says which column use for the new column names (Torgersen, Dream, Biscoe)
values_from = says which column to pull the values for those new columns from
Grouped operations
In Stata, the by command is used as a prefix for certain functions
The data must be sorted before the by operation can be used.
Grouping can be done in two locations.
As part of a pipe:
Or as part of a function, if it permits:
Miscellaneous operations
Seeds
Automating tables
Tabulation options
There are a few good options for making and formatting tables in R:
gtgtsummarykable(withkableExtra)tinytableflextable
Depending on your output, some packages work better than others. For things like HTML and PDF, they all work fairly well. For Word output (docx), gt doesn’t do so well.
Data visualisation
Analysing data
Reproducible documents
The situation
You are drafting a report or paper using Microsoft Word.
You have:
- Four paragraphs with in-text reporting of results
- Three tables
- Two figures
- And a partridge in a pear tree
The problem
Oh no! You now have to update everything because of:
- Changes in the data
- Changes in the analysis
- Changes due to reviewer comments
- Changes in the wind
The solution
Use Quarto to write your report/paper and automatically generate the tables, figures, and in-text reporting!
This has several benefits:
- Changes to data are automatically incorporated, everywhere
- Human error is limited to the initial setup, not every time data needs updating
- Updating takes as little as 5 seconds
- You learn a cool new skill
What is Quarto?
Quarto is open-source scientific and technical publishing system. It lets you:
- Write documents, presentations, and more
- Output these documents in Word and PDF
- Format text explicitly using markup
- Run code within the same document (more on this later)
- Manage citations, styles, table and figure numbering
- Most importantly, it makes your work more reproducible!
What does Quarto look like?
Text written with Quarto uses markup to apply styling. In this sentence, the word **bold** is bolded and *italic* is italicised.
{fig-align="left" width=20%}
You can do all the normal stuff:
1. Make lists
2. Add citations
3. etc.Text written with Quarto uses markup to apply styling. In this sentence, the word bold is bolded and italic is italicised.

You can do all the normal stuff:
- Make lists
- Add citations
- etc.
What would a workflow look like?
You can create workflow that suits you. I can imagine people:
- Doing their analysis in Stata
- Creating figures using Stata
- Exporting results into an Excel spreadsheet
- Reading the spreadsheet into R
- Using Quarto for write-up
- Data cleaning with Stata
- Saving analysis datasets
- Importing these datasets into R/Quarto
- Creating figures and tables with R
- Writing up with Quarto
- Doing everything with R/Quarto
A real example
I recently had a paper published: https://doi.org/10.1016/j.anzjph.2025.100249
I did everything we’ve just covered when writing up this paper:
- Wrote the whole paper
- Automated in-text numbers
- Created tables
- Included figures created elsewhere
- Automated table/figure numbering
- Referencing
My entire thesis is like this actually — the code and files are available: https://github.com/benharrap/thesis
A real example
Every Quarto document starts with the parameters:
Then the files get read in
A real example
| Demographic and other characteristics for the matched cohort of never- and ever-placed children | ||
|---|---|---|
| Never-placed | Ever-placed | |
| N | 7,442 | 3,721 |
| IRSAD at birth | ||
| 1st quintile | 3,559 (47.8%) | 1,781 (47.9%) |
| 2nd quintile | 1,336 (18.0%) | 675 (18.1%) |
| 3rd quintile | 1,717 (23.1%) | 836 (22.5%) |
| 4th quintile | 655 (8.8%) | 334 (9.0%) |
| 5th quintile | 175 (2.4%) | 95 (2.6%) |
| Remoteness area at birth | ||
| Major cities | 3,802 (51.1%) | 1,899 (51.0%) |
| Inner regional | 368 (4.9%) | 189 (5.1%) |
| Outer regional | 1,101 (14.8%) | 552 (14.8%) |
| Remote | 1,371 (18.4%) | 671 (18.0%) |
| Very remote | 800 (10.7%) | 410 (11.0%) |
| All hospital periods | ||
| All ages | 43,962 | 29,369 |
| Ages 0-4 | 22,048 (50.2%) | 15,586 (53.1%) |
| Ages 5-9 | 12,997 (29.6%) | 7,576 (25.8%) |
| Ages 10-14 | 8,913 (20.3%) | 6,207 (21.1%) |
| All PPH periods | ||
| All ages | 7,630 | 6,491 |
| Ages 0-4 | 5,366 (70.3%) | 4,850 (74.7%) |
| Ages 5-9 | 1,752 (23%) | 1,227 (18.9%) |
| Ages 10-14 | 512 (6.7%) | 414 (6.4%) |
A real example
While government departments in Australia [@aihw2020b;@dohwa2017;@ahmac2017] and researchers [@li2009;@guthrie2012] report rates of PPHs...
After excluding `r n_missing` who had missing data on the matching variables, a total of `r n_matching` children were included in the matching process. There were `r n_ever` ever-placed children who were matched with `r n_never` never-placed children.
When considering the mechanisms for prevention suggested by Anderson et al. [@anderson2012], most conditions were preventable through early access to primary care.
While government departments in Australia1–3 and researchers4,5 report rates of PPHs…
After excluding 125 who had missing data on the matching variables, a total of 33,403 children were included in the matching process. There were 3,721 ever-placed children who were matched with 7,442 never-placed children.
When considering the mechanisms for prevention suggested by Anderson et al.6, most conditions were preventable through early access to primary care.
References

Slides available from https://benharrap.github.io/workshops/2025-02-14-coding-in-stata-and-r/slides.html